 ] to go to the
graphical result
] to go to the
graphical result
Click on [ ] to go to the
graphical result
] to go to the
graphical result
Warning: Original output has been filtered to yield non-redundant similarities BLASTP 2.1.3 [Apr-1-2001] Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= D (1051 letters) Database: ProDom2001.1 multiple alignments 1,057,487 sequences; 119,872,694 total letters Searching..................................................done ProDom domains producing High-scoring Segment Pairs: Position ProDom domain Score E value 93-238 #PD315907 FH F21O3.25 GB|AAF02158.1 NFH1 387 3e-36 191-264 #PD320590 NFH1 127 5e-06 239-581 #PD320567 AHF1 FORMIN-LIKE 914 3e-97 582-699 #PD225067 FORMIN-LIKE FH P140MDIA AHF1 571 2e-57 594-709 #PD004159 COILED DIAPHANOUS HOMOLOG LD 100 0.007 710-842 #PD003542 CHROMOSOME COILED CELL DIAPHANOUS 586 3e-59 844-922 #PD331679 FORMIN-LIKE AHF1 NFH2 F18O19.9 368 6e-34 923-1012 #PD015702 FORMIN-LIKE FH F21O3.25 AHF1 349 8e-32 >PD320567 (Closest domain: Q9SE97_ARATH 239-581) Number of sequences in family: 1 Most frequent protein names: Commentary (automatic): AHF1 FORMIN-LIKE Length = 343 Score = 914 (356 bits), Expect = 3e-97 Identities = 224/343 (65%), Positives = 224/343 (65%) Query: 239 PDVGSIGEEDEEDEFYSPRGSQSGREPLNRVGLPGQNPRXXXXXXXXXXXXXXXXXXXXX 298 PDVGSIGEEDEEDEFYSPRGSQSGREPLNRVGLPGQNPR Sbjct: 239 PDVGSIGEEDEEDEFYSPRGSQSGREPLNRVGLPGQNPRSVNNDTISCSSSSSGSPGRST 298 Query: 299 XXXISPSMSPKRSEPKPPVISTPEPAELTDYRFVRXXXXXXXXXXXXXXXXDEVGLNQIF 358 ISPSMSPKRSEPKPPVISTPEPAELTDYRFVR DEVGLNQIF Sbjct: 299 FISISPSMSPKRSEPKPPVISTPEPAELTDYRFVRSPSLSLASLSSGLKNSDEVGLNQIF 358 Query: 359 RSPTVTSLTTSPENNKKENSPLSSTSTSPERRPNDTPEAYLRXXXXXXXXXXPYRCFQKS 418 RSPTVTSLTTSPENNKKENSPLSSTSTSPERRPNDTPEAYLR PYRCFQKS Sbjct: 359 RSPTVTSLTTSPENNKKENSPLSSTSTSPERRPNDTPEAYLRSPSHSSASTSPYRCFQKS 418 Query: 419 PEVLPAFMXXXXXXXXXXXXXXXXXXHGGQGFLKQLDALXXXXXXXXXXXXXXXXEKASH 478 PEVLPAFM HGGQGFLKQLDAL EKASH Sbjct: 419 PEVLPAFMSNLRQGLQSQLLSSPSNSHGGQGFLKQLDALRSRSPSSSSSSVCSSPEKASH 478 Query: 479 KSPVTXXXXXXXXXXXXXXXXXXXXXXXLDVSPRISNISPQILQSRVXXXXXXXXXXXXW 538 KSPVT LDVSPRISNISPQILQSRV W Sbjct: 479 KSPVTSPKLSSRNSQSLSSSPDRDFSHSLDVSPRISNISPQILQSRVPPPPPPPPPLPLW 538 Query: 539 GRRSQVTTKADTISRPPSLTPPSHPFVIPSENLPVTSSPMETP 581 GRRSQVTTKADTISRPPSLTPPSHPFVIPSENLPVTSSPMETP Sbjct: 539 GRRSQVTTKADTISRPPSLTPPSHPFVIPSENLPVTSSPMETP 581 >PD003542 (Closest domain: Q9SE97_ARATH 710-842) Number of sequences in family: 59 Most frequent protein names: DIA1(2) Commentary (automatic): CHROMOSOME COILED CELL DIAPHANOUS FORMIN SPLICING DEVELOPMENTAL HOMOLOG ALTERNATIVE Length = 133 Score = 586 (230 bits), Expect = 3e-59 Identities = 121/133 (90%), Positives = 121/133 (90%) Query: 710 MAPTKEEERKLKAYNDDSPVKLGHAEKFLKAMLDIPFAFKRVDAMLYVANFESEVEYLKK 769 MAPTKEEERKLKAYNDDSPVKLGHAEKFLKAMLDIPFAFKRVDAMLYVANFESEVEYLKK Sbjct: 710 MAPTKEEERKLKAYNDDSPVKLGHAEKFLKAMLDIPFAFKRVDAMLYVANFESEVEYLKK 769 Query: 770 SFETLEAACEELRNSRMFLKLLEAVLKTGNRMNVGTNRGDAHAFXXXXXXXXXXXXGADG 829 SFETLEAACEELRNSRMFLKLLEAVLKTGNRMNVGTNRGDAHAF GADG Sbjct: 770 SFETLEAACEELRNSRMFLKLLEAVLKTGNRMNVGTNRGDAHAFKLDTLLKLVDVKGADG 829 Query: 830 KTTLLHFVVQEII 842 KTTLLHFVVQEII Sbjct: 830 KTTLLHFVVQEII 842 >PD225067 (Closest domain: Q9SE97_ARATH 582-699) Number of sequences in family: 13 Most frequent protein names: Commentary (automatic): FORMIN-LIKE FH P140MDIA AHF1 F23H11.22 GB|AAF02158.1 CHROMOSOME F21O3.25 Length = 118 Score = 571 (224 bits), Expect = 2e-57 Identities = 118/118 (100%), Positives = 118/118 (100%) Query: 582 ETVCASEAAEETPKPKLKALHWDKVRASSDREMVWDHLRSSSFKLDEEMIETLFVAKSLN 641 ETVCASEAAEETPKPKLKALHWDKVRASSDREMVWDHLRSSSFKLDEEMIETLFVAKSLN Sbjct: 582 ETVCASEAAEETPKPKLKALHWDKVRASSDREMVWDHLRSSSFKLDEEMIETLFVAKSLN 641 Query: 642 NKPNQSQTTPRCVLPSPNQENRVLDPKKAQNIAILLRALNVTIEEVCEALLEGNADTL 699 NKPNQSQTTPRCVLPSPNQENRVLDPKKAQNIAILLRALNVTIEEVCEALLEGNADTL Sbjct: 642 NKPNQSQTTPRCVLPSPNQENRVLDPKKAQNIAILLRALNVTIEEVCEALLEGNADTL 699 >PD315907 (Closest domain: Q9SE97_ARATH 89-238) Number of sequences in family: 6 Most frequent protein names: Commentary (automatic): FH F21O3.25 GB|AAF02158.1 NFH1 FORMIN-LIKE NFH2 AHF1 F18O19.9 Length = 150 Score = 387 (153 bits), Expect = 3e-36 Identities = 89/146 (60%), Positives = 89/146 (60%) Query: 93 LIVPHATKSPPNSKKXXXXXXXXXXXXXXXXXXXXXXYWRRSKRNQDLNFXXXXXXXXXX 152 LIVPHATKSPPNSKK YWRRSKRNQDLNF Sbjct: 93 LIVPHATKSPPNSKKLLIVAISAVSSAALVALLIALLYWRRSKRNQDLNFSDDSKTYTTD 152 Query: 153 XXRRVYXXXXXXXXXXXXNAEARSKQRXXXXXXXXXXXEFLYLGTMVNQRGIDEQSLSNN 212 RRVY NAEARSKQR EFLYLGTMVNQRGIDEQSLSNN Sbjct: 153 SSRRVYPPPPATAPPTRRNAEARSKQRTTTSSTNNNSSEFLYLGTMVNQRGIDEQSLSNN 212 Query: 213 GSSSRKLESPDLQPLPPLMKRSFRLN 238 GSSSRKLESPDLQPLPPLMKRSFRLN Sbjct: 213 GSSSRKLESPDLQPLPPLMKRSFRLN 238 >PD331679 (Closest domain: Q9SE97_ARATH 844-922) Number of sequences in family: 4 Most frequent protein names: Commentary (automatic): FORMIN-LIKE AHF1 NFH2 F18O19.9 Length = 79 Score = 368 (146 bits), Expect = 6e-34 Identities = 79/79 (100%), Positives = 79/79 (100%) Query: 844 AEGTRLSGNNTQTDDIKCRKLGLQVVSSLCSELSNVKKAAAMDSEVLSSYVSKLSQGIAK 903 AEGTRLSGNNTQTDDIKCRKLGLQVVSSLCSELSNVKKAAAMDSEVLSSYVSKLSQGIAK Sbjct: 844 AEGTRLSGNNTQTDDIKCRKLGLQVVSSLCSELSNVKKAAAMDSEVLSSYVSKLSQGIAK 903 Query: 904 INEAIQVQSTITEESNSQR 922 INEAIQVQSTITEESNSQR Sbjct: 904 INEAIQVQSTITEESNSQR 922 >PD015702 (Closest domain: Q9SDN5_TOBAC 721-814) Number of sequences in family: 13 Most frequent protein names: Commentary (automatic): FORMIN-LIKE FH F21O3.25 AHF1 F23H11.22 GB|AAF02158.1 CHROMOSOME NFH1 F22F7.8 Length = 94 Score = 349 (138 bits), Expect = 8e-32 Identities = 68/90 (75%), Positives = 82/90 (90%) Query: 923 FSESMKTFLKRAEEEIIRVQAQESVALSLVKEITEYFHGNSAKEEAHPFRIFLVVRDFLG 982 FSESM F+K AEE+I+R+QAQE++A+SLVKEITEY HG+SA+EEAHPFRIF+VV+DFL Sbjct: 721 FSESMNRFMKVAEEKILRLQAQETLAMSLVKEITEYVHGDSAREEAHPFRIFMVVKDFLM 780 Query: 983 VVDRVCKEVGMINERTMVSSAHKFPVPVNP 1012 ++D VCKEVG INERT+VSSA KFPVPVNP Sbjct: 781 ILDCVCKEVGTINERTIVSSAQKFPVPVNP 810 >PD320590 (Closest domain: Q9SDN6_TOBAC 176-518) Number of sequences in family: 1 Most frequent protein names: Commentary (automatic): NFH1 Length = 343 Score = 127 (53.3 bits), Expect = 5e-06 Identities = 43/94 (45%), Positives = 56/94 (58%), Gaps = 21/94 (22%) Query: 191 EFLYLGTMVNQR-GID------EQSLSNNGSS---SRKLESPDLQPLPPLMKRSFR---- 236 EFLYLGTMV+ GID ++ S N +S SRK++SP++ PLPPL+ R+ Sbjct: 189 EFLYLGTMVSSHGGIDGPHNPPQRRRSGNVTSVPASRKMDSPEIHPLPPLLGRNLSQNYG 248 Query: 237 ------LNPDVGSIGEEDEEDEFYSPRGSQSGRE 264 N DV S G +E++EFYSPRGS GRE Sbjct: 249 NNNDNNNNADVIS-GRTEEDEEFYSPRGSLDGRE 281 >PD004159 (Closest domain: DIA2_HUMAN 622-754) Number of sequences in family: 9 Most frequent protein names: DIA1(2) Commentary (automatic): COILED DIAPHANOUS HOMOLOG LD CELL P140MDIA SIMILAR CHICKEN PIR:A41724 Length = 133 Score = 100 (42.9 bits), Expect = 0.007 Identities = 31/120 (25%), Positives = 59/120 (48%), Gaps = 4/120 (3%) Query: 594 PKPKLKALHWDKVRASSDRE-MVWDHLRSSSFKLDEEMIE-TLFVAKSLNNKPNQSQTTP 651 P+ +K ++W K+ + E W ++ F+ + + L A + + N Sbjct: 634 PEVSMKRINWSKIEPTELSENCFWLRVKEDKFENPDLFAKLALNFATQIKVQKNAEALEE 693 Query: 652 RCVLPSPN--QENRVLDPKKAQNIAILLRALNVTIEEVCEALLEGNADTLGTELLESLLK 709 + P+ +E R+LDPK AQN++I L + + E++ +LE N D L L+++L+K Sbjct: 694 KKTGPTKKKVKELRILDPKTAQNLSIFLGSYRMPYEDIRNVILEVNEDMLSEALIQNLVK 753 Parameters: Database: ProDom2001.1 multiple alignments Number of letters in database: 119,872,694 Number of sequences in database: 1,057,487 Lambda K H 0.313 0.130 0.364 Lambda K H 0.267 0.0410 0.140 Matrix: BLOSUM62 Gap Penalties: Existence: 11, Extension: 1 Number of Hits to DB: 171945910 Number of Sequences: 1057487 Number of extensions: 5470952 Number of successful extensions: 16217 Number of sequences better than 1.0e-02: 82 Number of HSP's better than 0.0 without gapping: 59 Number of HSP's successfully gapped in prelim test: 23 Number of HSP's that attempted gapping in prelim test: 16109 Number of HSP's gapped (non-prelim): 85 length of query: 1051 length of database: 119,872,694 effective HSP length: 60 effective length of query: 991 effective length of database: 56,423,474 effective search space: 55915662734 effective search space used: 55915662734 T: 11 A: 40 X1: 16 ( 7.2 bits) X2: 38 (14.6 bits) X3: 64 (24.7 bits) S1: 42 (21.9 bits) S2: 98 (42.4 bits)
Clickable ncbi-blastp2 output (Go to HSPs)
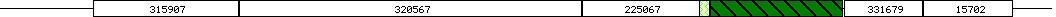
Domain ID Sequence BEGIN Sequence END
